A. Baumannii MLA Complex
Dataset
Learning objectives
Importing results into CryoSPARC, 3D classification techniques. Note: Use the outputs of the previous step as the inputs for the next step, unless otherwise noted.
| Title | Value |
|---|---|
| Title | |
| Description | Leave empty |
| Parameter | Value |
|---|---|
| Particle meta path |
| Parameter | Value |
|---|---|
| Fourier crop to box size (pix) |
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize |
Choose a set of 2D classes (approximately 20) that look sharp and display secondary structure features.
You should end up with around 35-40K particles.
Use default parameters
--- Queue the following two refinement jobs in parallel. ---
| Parameter | Value |
|---|---|
| Symmetry |
| Parameter | Value |
|---|---|
| Symmetry |
- Download the refined volume from the previous Non-uniform Refinement job.
-
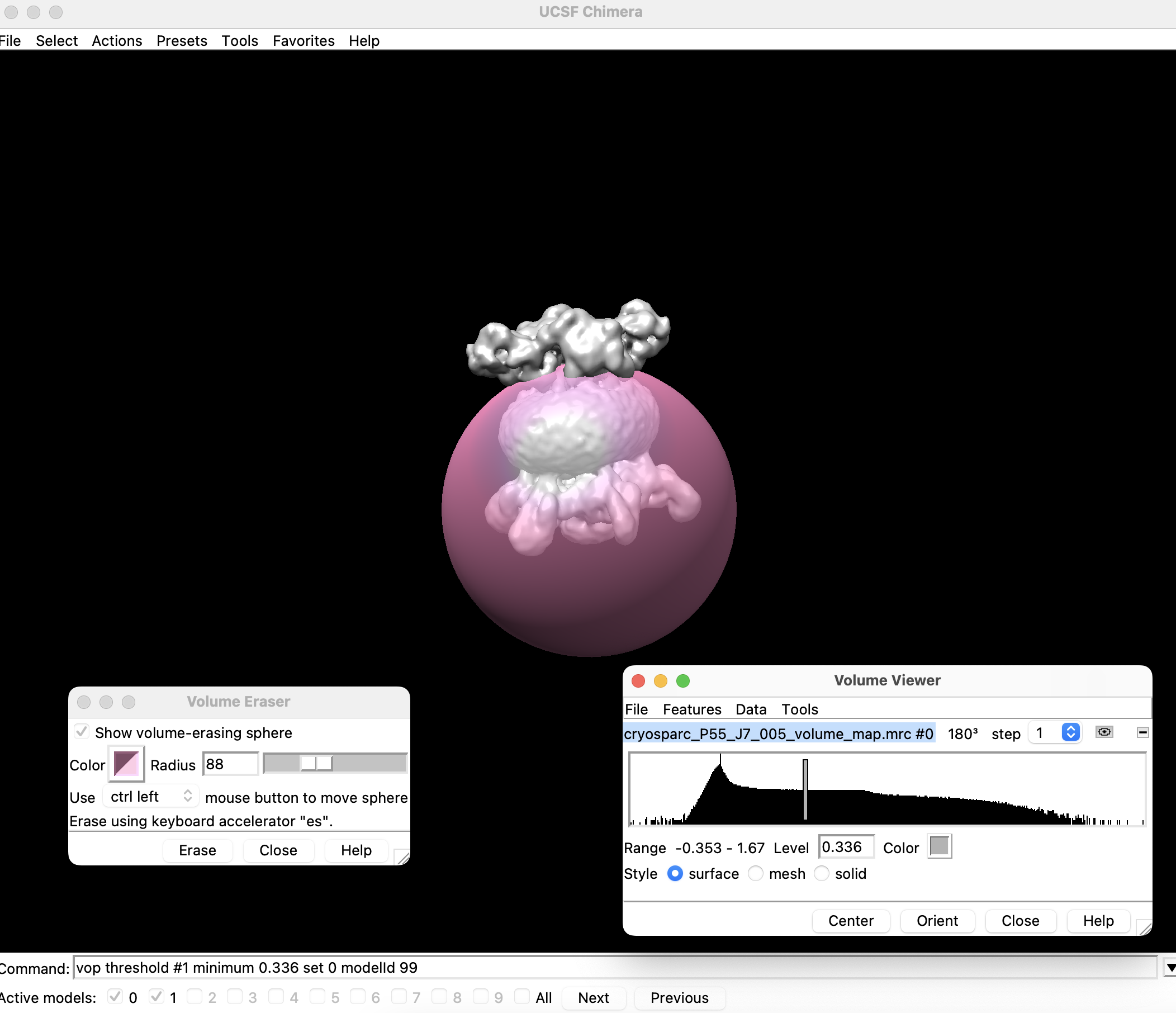
Open the map in UCSF Chimera, and note the opened structures model ID as
<model_id>. -
Decrease the threshold until you can clearly see both "lobes" (these are the MlaB domains, located opposite from the 6-fold symmetric MlaD domain). Note the threshold value
<threshold_number>. - Open the Map Eraser (Under Tools > Volume Data > Volume Eraser ).
- Erase the micelle and the 6-fold symmetric MlaD domain.
-
Use
to binarize the density map of the MlaB domain, to convert it to a mask.
- Set all density values outside of the MlaB domain to 0:
- Set all density values within the MlaB domain to 1:
-
Save the mask to your home directory. In the Volume Viewer, select File -> Save map as ->
baum_embolab_mask.mrc
| Parameter | Value |
|---|---|
| Volume data path | |
| Type of volume being imported |
| Parameter | Value |
|---|---|
| Type of input volume | |
| Type of output volume | |
| Threshold (must set to process mask) | |
| Dilation radius | |
| Soft padding width |
--- Queue 3D classification and 3DVA in parallel. ---
| Input | Source |
|---|---|
| Particle stacks | Particles from Non-uniform Refinement |
| Initial volumes | Leave empty |
| Solvent mask | Mask from Non-uniform Refinement |
| Focus mask | Mask from Volume Tools |
| Parameter | Value |
|---|---|
| Number of classes | |
| Number of O-EM epochs |
| Input | Source |
|---|---|
| Particle stacks | Particles from Non-uniform Refinement |
| Initial volumes | Leave empty |
| Solvent mask | Mask from Non-uniform Refinement |
| Focus mask | Mask from Volume Tools |
| Parameter | Value |
|---|---|
| Number of classes | |
| Number of O-EM epochs | |
| Force hard classification |
| Input | Source |
|---|---|
| Particle stacks | Particles from Non-uniform Refinement |
| Mask | Mask from Non-uniform Refinement |
| Parameter | Value |
|---|---|
| Filter resolution (A) | |
| Downsample to box size |
| Parameter | Value |
|---|---|
| Downsample to box size |
| Parameter | Value |
|---|---|
| Output mode | |
| Number of frames | |
| Downsample to box size | |
| Intermediates: window (frames) |
